Indices and tables¶
Doxygen index test¶
- struct
- struct
- struct
Public Functions
-
simulation::results::results(size_t _N)¶
Public Members
-
std::unique_ptr<double[]>
simulation::results::mx¶
-
std::unique_ptr<double[]>
simulation::results::my¶
-
std::unique_ptr<double[]>
simulation::results::mz¶
-
std::unique_ptr<double[]>
simulation::results::field¶
-
std::unique_ptr<double[]>
simulation::results::time¶
-
size_t
simulation::results::N¶
-
- class
- #include <rng.hpp>
Abstract class for random number generators.
Subclassed by RngArray, RngMtDownsample, RngMtNorm
Public Functions
-
virtual double
Rng::get()¶
= 0 Get a single random number.
- Return
- a single draw from the random number generator
-
virtual double
- class
- #include <rng.hpp>
Provides an
Rnginterface to a predefined array of numbers.Inherits from Rng
Public Functions
-
RngArray::RngArray(const double *arr, size_t arr_length, size_t stride = 1)¶ Default constructor for RngArray.
- Parameters
_arr: a predefined array of random numbers_arr_length: length of the predefined array_stride: the number of consecutive elements to stride for each call to.get()
-
double
RngArray::get()¶ Get the next (possibly stridden) value from the array.
The first call will always return the value at the 0th index. Each subsequent call will stride the array (default value is 1) and return that value. A call that extends beyond the end of the array will result in an error.
- Exceptions
std::out_of_range: when a call attempts to get a value beyond the maximum length of the predefined array.
-
- class
- #include <rng.hpp>
Generate normally distributed values with downsampling.
Uses the Mersenne Twister to generate normally distributed random numbers. Down-samples the stream of random numbers by summing consecutive draws along each dimension. Function is usually used for generating coarse Wiener processes
Inherits from Rng
Public Functions
-
RngMtDownsample::RngMtDownsample(const unsigned long int seed, const double std, const size_t dim, const size_t down_factor)¶ Default constructor for RngMtDownsample.
-
double
RngMtDownsample::get()¶ Get a single downsampled value from the random number generator.
Private Functions
-
void
RngMtDownsample::downsample_draw()¶
Private Members
-
std::mt19937_64
RngMtDownsample::generator¶ A Mersenne Twister generator instance.
-
std::normal_distribution<double>
RngMtDownsample::dist¶ A normal distribution instance.
-
int
RngMtDownsample::current_dim¶ Stores the current state of the output dimenstion.
-
std::vector<double>
RngMtDownsample::store¶ Stores consecutive random numbers.
-
const size_t
RngMtDownsample::D¶ The number of dimensions required.
-
const size_t
RngMtDownsample::F¶ The number of consecutive random numbers to downsample.
-
- class
- #include <rng.hpp>
Uses Mersenne Twister to generate normally distributed values.
Random numbers have a mean of zero and a user specified standard deviation.
Inherits from Rng
Public Functions
-
RngMtNorm::RngMtNorm(const unsigned long int seed, const double std)¶ Default constructor for RngMtNorm.
- Parameters
seed: seed for random number generatorstd: the standard deviation of the normally distributed numbers
-
double
RngMtNorm::get()¶ Draw a single normally distributed value from the RNG.
- Return
- single normally distributed number
-
-
namespace
conf¶ Variables
-
list conf.extensions
-
dictionary conf.breathe_projects
-
string conf.breathe_default_project
-
list conf.templates_path
-
string conf.source_suffix
-
string conf.master_doc
-
string conf.project
-
string conf.copyright
-
string conf.author
-
string conf.version
-
string conf.release
-
language¶
-
list conf.exclude_patterns
-
string conf.pygments_style
-
todo_include_todos¶
-
string conf.html_theme
-
list conf.html_static_path
-
string conf.htmlhelp_basename
-
dictionary conf.latex_elements
-
list conf.latex_documents
-
list conf.man_pages
-
list conf.texinfo_documents
-
tuple conf.read_the_docs_build
-
string conf.SUFFIX
-
using_rtd_theme¶
-
html_style¶
-
dictionary conf.html_theme_options
-
list conf.html_theme_path
-
string conf.websupport2_base_url
-
string conf.websupport2_static_url
-
dictionary conf.context
-
html_context¶
-
- namespace
-
namespace
convergence¶ Variables
-
tuple convergence.dt
-
tuple convergence.convergence_data
-
tuple convergence.diffs
-
tuple convergence.ensemble_diff
-
tuple convergence.A
-
tuple convergence.b
-
tuple convergence.least_square_res
-
list convergence.rate
-
dictionary convergence.results
-
- namespace
Contains higher order functions for currying.
Functions
- template <typename RET, typename FIRST>
-
std::function<RET()>
curry::curry(std::function<RET(FIRST)> f, FIRST x, )¶ Curry single argument function.
Binds a function’s single argument and returns the callable function without arguments.
- Return
- std::function with no arguments
- Parameters
f: std::function with a single argumentx: value of argument to bind to function
- template <typename RET, typename FIRST, typename... REST>
-
std::function<RET()>
curry::curry(std::function<RET(FIRST, REST...)> f, FIRST x, REST... rest, )¶ Curry multi-argument function.
Binds the multiple arguments to a std::function’s signature and returns the call without function call with no arguments
- Return
- std::function with no arguments
- Parameters
f: std::function with multiple argumentsx: value of first argument to bind to functionrest...: variable number of arguments to bind (can have any type)
- template <typename RET, typename FIRST, typename... ARGS>
-
std::vector<std::function<RET()>>
curry::vector_curry(std::function<RET(FIRST, ARGS...)> f, std::vector<FIRST> first, std::vector<ARGS>... args, )¶ Curry multi-argument function many times with lists of arguments.
Maps multiple vectors of function arguments to a vector of callable functions with all parameters bound. For example given the add function
returns x+yand two std::vector<double> instances withxs={1,2},ys={3,4}then the vector_curry will return astd::vector<std::function<double()> >with two items. The first call will evaluate to1+3=4and a call to the second item will evaluate to2+4=6.- Return
- std::vector of callable functions
- Parameters
f: std::function with multiple argumentsfirst: std::vector of different values for first argumentargs...: variable number of std::vectors with different values of function arguments (all vectors must be same length)
- namespace
discrete orientation model for magnetic particles
Functions
-
void
dom::transition_matrix(double *W, const double k, const double v, const double T, const double h, const double tau0)¶ Compute the 2x2 transition matrix for a single particle.
Assumes uniaxial anisotropy and an applied field h<1 Only valid for large energy barriers
-
void
dom::master_equation_with_update(double *derivs, double *work, const double k, const double v, const double T, const double tau0, const double t, const double *state_probabilities, const std::function<double(double)> applied_field)¶ Computes master equation for particle in time-dependent field.
Computes the applied field in the z-direction at time t. Uses the field value to compute the transition matrix and corresponding master equation derivatives for the single particle.
- Parameters
derivs: master equation derivatives [length 2]work: vector [length 4]k: anisotropy strength constant for the uniaxial anisotropyv: volume of the particle in meter^3T: temperature of environment in Kelvintau0: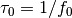 where f0 is the attempt
where f0 is the attempt t: time at which to evaluate the external fieldstate_probabilities: the current state probabilities for each of the 2 states (up and down) [length 2]applied_field: a scalar function takes a double and returns a double. Given the current time should return normalised field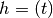 where the field is normalised by
where the field is normalised by 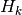
-
void
- namespace
Functions
-
void
driver::rk4(double *states, const double *initial_state, const std::function<void(double *, const double *, const double)> derivs, const size_t n_steps, const size_t n_dims, const double step_size, )¶
-
void
driver::eulerm(double *states, const double *initial_state, const double *wiener_process, const std::function<void(double *, const double *, const double)> drift, const std::function<void(double *, const double *, const double)> diffusion, const size_t n_steps, const size_t n_dims, const size_t n_wiener, const double step_size, )¶
-
void
driver::heun(double *states, const double *initial_state, const double *wiener_process, const std::function<void(double *, const double *, const double)> drift, const std::function<void(double *, const double *, const double)> diffusion, const size_t n_steps, const size_t n_dims, const size_t n_wiener, const double step_size, )¶
-
void
-
namespace
equilibrium¶ -
Variables
-
tuple equilibrium.mz
-
tuple equilibrium.theta
-
tuple equilibrium.config
-
tuple equilibrium.t
-
tuple equilibrium.analytic
-
- namespace
Contains functions for computing magnetic fields.
Functions for computing effective fields from anistoropy and a range of time-varying applied fields.
- Author
- Oliver Laslett
- Date
- 2017
Functions
-
double
field::constant(const double h, const double t)¶ A constant applied field.
A simple placeholder function representing a constant field. Always returns the same value.
- Return
- the constant field amplitude at all values of time
- Parameters
h: applied field amplitudet: time (parameter has no effect)
-
double
field::sinusoidal(const double h, const double f, const double t)¶ A sinusoidal alternating applied field.
Returns the value of a sinusoidally varying field at any given time.
- Return
- the value of the varying applied field at time
t - Parameters
h: applied field amplitudef: applied field frequencyt: time
-
double
field::square(const double h, const double f, const double t)¶ A square wave switching applied field.
An alternating applied field with a square shape centred around zero. i.e. it alternates between values
-handh- Return
- the value of the square wave applied field at time
t - Parameters
h: applied field amplitudef: applied field frequencyt: time
-
double
field::square_fourier(const double h, const double f, const size_t n_compononents, double t)¶ A square wave applied field of finite Fourier components.
An approximate square wave is computed from a finite number of Fourier components. The square wave alternates between
-handh.- Return
- the value of the square wave applied field at time
t - Parameters
h: applied field amplitudef: applied field frequencyn_components: number of Fourier components to computet: time
-
void
field::uniaxial_anisotropy(double *h_anis, const double *magnetisation, const double *anis_axis)¶ Effective field contribution from uniaxial anisotropy.
The effective field experienced by a single particle with a uniaxial anisotropy.
- Parameters
h_anis: effective field [length 3]mag: the magnetisation of the particle of [length 3]axis: the anisotropy axis of the particle [length 3]
-
void
field::uniaxial_anisotropy_jacobian(double *jac, const double *axis)¶ Jacobian of the uniaxial anisotropy effective field term.
The Jacobian of a particle’s uniaxial anisotropy with respect to it’s magnetisation value.
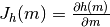
- Parameters
jac: the jacobian of the effective field [length 3*3]axis: the anisotropy axis of the particle [length 3]
- namespace
Functions
-
void
integrator::rk4(double *next_state, double *k1, double *k2, double *k3, double *k4, const double *current_state, const std::function<void(double *, const double *, const double)> derivs, const size_t n_dims, const double t, const double step_size, )¶
-
void
integrator::heun(double *next_state, double *drift_arr, double *trial_drift_arr, double *diffusion_matrix, double *trial_diffusion_matrix, const double *current_state, const double *wiener_steps, const std::function<void(double *, const double *, const double)> drift, const std::function<void(double *, const double *, const double)> diffusion, const size_t n_dims, const size_t wiener_dims, const double t, const double step_size, )¶
-
void
integrator::eulerm(double *states, double *diffusion_matrix, const double *initial_state, const double *wiener_process, const std::function<void(double *, const double *, const double)> drift, const std::function<void(double *, const double *, const double)> diffusion, const size_t n_dims, const size_t n_wiener, const double t, const double step_size, )¶
- template <class CSTATES, class CDIFF>
-
void
integrator::milstein(CSTATES &next_state, const CSTATES ¤t_state, const CSTATES &drift, const CDIFF &diffusion, const CSTATES &wiener_increments, const double step_size)¶
-
int
integrator::implicit_midpoint(double *x, double *dwm, double *a_work, double *b_work, double *adash_work, double *bdash_work, double *x_guess, double *x_opt_tmp, double *x_opt_jac, lapack_int *x_opt_ipiv, const double *x0, const double *dw, const std::function<void(double *, double *, double *, double *, const double *, const double, const double)> sde, const size_t n_dim, const size_t w_dim, const double t, const double dt, const double eps, const size_t max_iter, )¶
-
void
- namespace
Functions
- template <typename T>
-
int
io::write_array(const std::string fname, T const *arr, const size_t len)¶
- namespace
Functions for evaluating the Landau-Lifshitz-Gilbert equation.
Includes the basic equation as well as Jacobians and combined functions to update fields during integration.
- Author
- Oliver Laslett
- Date
- 2017
Functions
-
void
llg::drift(double *deriv, const double *state, const double time, const double alpha, const double *heff)¶ Deterministic drift component of the stochastic LLG.
- Parameters
deriv: drift derivative of the deterministic part of the stochastic llg [length 3]state: current state of the magnetisation vector [length 3]t: time (has no effect)alpha: damping ratiothe: effective field on the magnetisation [length 3]
-
void
llg::drift_jacobian(double *deriv, const double *state, const double time, const double alpha, const double *heff, const double *heff_jac)¶ Jacobian of the deterministic drift component of the stochastic LLG Since, in the general case, the effective field is a function of the magnetisation, the Jacobian of the effective field must be known in order to compute the Jacobian of the drift component.
- Parameters
jac: Jacobian of the drift [length 3x3]m: state of the magnetisation vector [length 3]t: time (has no effect)a: damping ratio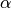
h: effective field acting on the magnetisation [length 3]hj: Jacobian of the effective field evaluated at the current value ofm[length 3x3]
-
void
llg::diffusion(double *deriv, const double *state, const double time, const double sr, const double alpha)¶ The stochastic diffusion component of the stochastic LLG.
- Parameters
deriv: diffusion derivatives [length 3x3]state: current state of the magnetisation [length 3]t: time (has no effect)sr: normalised noise power of the thermal field (see notes on LLG normalisation for details)alpha: damping ratio
-
void
llg::diffusion_jacobian(double *jacobian, const double *state, const double time, const double sr, const double alpha)¶ Jacobian of the stochastic diffusion component of the LLG.
- Parameters
jacobian: Jacobian of the LLG diffusion [length 3x3]state: current state of the magnetisation vector [length 3]t: time (has no effect)sr: normalised noise power of the thermal field (see notes on LLG normalisation for details)alpha: damping ratio
-
void
llg::sde_with_update(double *drift, double *diffusion, double *heff, const double *current_state, const double drift_time, const double diffusion_time, const double *happ, const double *anisotropy_axis, const double alpha, const double noise_power)¶ Computes drift and diffusion of LLG after updating the field.
The effective field is first computed based on the applied field and current state of the magnetisation. This is then used to compute the current drift and diffusion components of the LLG. Assumes uniaxial anisotropy.
- Parameters
drift: deterministic component of the LLG [length 3]diffusion: stochastic component of the LLG [length 3x3]heff: effective field including the applied field contribution [length 3]state: current state of the magnetisation [length 3]a_t: time at which to evaluate the driftb_t: time at which to evaluate the diffusionhapp: the applied field at timea_t[length 3]aaxis: the anisotropy axis of the particle [length 3]alpha: damping ratiosr: normalised noise power of the thermal field (see notes on LLG normalisation for details)
-
void
llg::jacobians_with_update(double *drift, double *diffusion, double *drift_jac, double *diffusion_jac, double *heff, double *heff_jac, const double *current_state, const double drift_time, const double diffusion_time, const double *happ, const double *anisotropy_axis, const double alpha, const double noise_power)¶ Computes effective field, drift, diffusion and Jacobians of LLG.
The effective field is first computed based on the applied field and current state of the magnetisation. This is then used to compute the drift, diffusion, and their respective Jacobians. Assumes uniaxial anisotropy.
- Parameters
drift: deterministic component of the LLG [length 3]diffusion: stochastic component of the LLG [length 3x3]drift_jac: Jacobian of the deterministic component [length 3x3]diffusion_jac: Jacobian of the diffusion component [length 3x3x3]heff: effective field including the applied field contribution [length 3]heff_jac: Jacobian of the effective field [length 3x3]state: current state of the magnetisation [length 3]a_t: time at which to evaluate the driftb_t: time at which to evaluate the diffusionhapp: the applied field at timea_t[length 3]aaxis: the anisotropy axis of the particle [length 3]alpha: damping ratios: normalised noise power of the thermal field (see notes on LLG normalisation for details)
- namespace
Typedefs
-
using
mnp::axis = typedef std::array<double,3>
-
using
- namespace
Functions
-
void
moma_config::validate_for_llg(const json input)¶ Validate that fields for llg simulation are in json.
Inspects a json file to check that it has the right structure and that parameters match assumptions required for simulation (e.g. temperature is not negative). Fatal logs on failure.
- Parameters
input: json config object
-
void
moma_config::validate_for_dom(const json input)¶ Validate that fields for discerete-orientation-model simulation are in json.
Inspects a json file to check that it has the right structure and that parameters match assumptions required for simulation (e.g. temperature is not negative). Fatal logs on failure.
- Parameters
input: json config object
-
void
moma_config::launch_simulation(const json input)¶ Main entry point for launching simulations from json config.
Runs simulations specified by the json configuration. The config must be normalised
moma_config::normalise. Function does not return anything but will write results to disk (see logs and config options).- Parameters
norm_config: json object of the normalised configuration file
-
void
- namespace
Functions
-
int
optimisation::newton_raphson_1(double *x_root, const std::function<double(const double)> f, const std::function<double(const double)> fdash, const double x0, const double eps = 1e-7, const size_t max_iter = 1000, )¶
-
int
optimisation::newton_raphson_noinv(double *x_root, double *x_tmp, double *jac_out, lapack_int *ipiv, int *lapack_err_code, const std::function<void(double *, double *, const double *)> func_and_jacobian, const double *x0, const lapack_int dim, const double eps = 1e-7, const size_t max_iter = 1000, )¶
-
int
- namespace
Functions
-
struct simulation::results
simulation::full_dynamics(const double damping, const double thermal_field_strength, const d3 anis_axis, const std::function<double(double)> applied_field, const d3 initial_magnetisation, const double time_step, const double end_time, Rng &rng, const bool renorm, const int max_samples, )¶
-
struct simulation::results
simulation::dom_ensemble_dynamics(const double volume, const double anisotropy, const double temperature, const double tau0, const std::function<double(double)> applied_field, const std::array<double, 2> initial_mags, const double time_step, const double end_time, const int max_samples, )¶ Simulates a single particle under the discrete orientation model.
Simulates a single uniaxial magnetic nanoparticle using the master equation. Assumes that the anisotropy axis is aligned with the external field in the $z$ direction.
The system is modelled as having two possible states (up and down). Given the initial probability that the system is in these two states, the master equation simulates the time-evolution of the probability of states. The magnetisation in the $z$ direction is computed as a function of the states.
- Return
- simulation::results struct containing the results of the simulation. The x and y components of the magnetisation are always zero.
- Parameters
damping: damping ratio - dimensionlessanisotropy: anisotropy constant - Kgm-3 ?temperature: temperature - Ktau0: reciprocal of the attempt frequency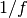 - s-1
- s-1 applied_field: a scalar in-out function that returns the value of the applied field in the z-direction at time tinitial_prbs: length 2 array of doubles with the initial probability of each state of the system.time_step: time_step for the integratorend_time: total length of the simulationmax_samples: integer number of times to sample the output. Setting to -1 will sample the output at every time step of the integrator.
- template <typename... T>
-
struct results
simulation::ensemble_run(const size_t max_samples, std::function<results(T...)> run_function, std::vector<T>... varying_arguments, )¶
- template <typename... T>
-
std::vector<d3>
simulation::ensemble_run_final_state(std::function<results(T...)> run_function, std::vector<T>... varing_arguments, )¶
- template <typename... T>
-
struct results
simulation::steady_state_cycle_dynamics(std::function<results(d3, T...)> run_function, const int max_samples, const double steady_state_condition, const std::vector<d3> initial_magnetisations, std::vector<T>... varying_arguments, )¶
-
struct simulation::results
- namespace
Functions
-
void
stochastic::strato_to_ito(double *ito_drift, const double *strato_drift, const double *diffusion, const double *diffusion_jacobian, const size_t n_dims, const size_t wiener_dims)¶
-
void
stochastic::ito_to_strato(double *strato_drift, const double *ito_drift, const double *diffusion, const double *diffusion_jacobian, const size_t n_dims, const size_t wiener_dims)¶
-
void
stochastic::master_equation(double *derivs, const double *transition_matrix, const double *current_state, const size_t dim)¶ Evaluates the derivatives of the master equation given a transition matrix.
The master equation is simply a linear system of ODEs, with coefficients described by the transition matrix.
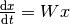
- Parameters
derivs: the master equation derivatives [length dim]transition_matrix: the [dim x dim] transition matrix (row-major)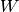
current_state: values of the state vector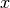
-
void
- namespace
Functions
-
double
trap::trapezoidal(double *x, double *y, size_t N)¶
-
double
-
file
conf.py
-
file
conf.py
-
file
constants.hpp
-
file
curry.hpp - #include <functional>#include <vector>#include “curry.tpp”
-
file
dom.hpp - #include <functional>
-
file
field.hpp - #include <cstddef>
-
file
integrators.hpp - #include <lapacke.h>#include <functional>
-
file
io.hpp - #include ”../include/easylogging++.h”#include <ctime>#include <cstring>#include <cstdlib>#include <cctype>#include <cwchar>#include <csignal>#include <cerrno>#include <cstdarg>#include <string>#include <vector>#include <map>#include <utility>#include <functional>#include <algorithm>#include <fstream>#include <iostream>#include <sstream>#include <memory>#include <type_traits>
-
file
llg.hpp
-
file
mnps.hpp - #include <array>
-
file
moma_config.hpp - #include <map>#include <string>#include “json.hpp”#include <algorithm>#include <array>#include <cassert>#include <ciso646>#include <cmath>#include <cstddef>#include <cstdint>#include <cstdlib>#include <cstring>#include <functional>#include <initializer_list>#include <iomanip>#include <iostream>#include <iterator>#include <limits>#include <locale>#include <memory>#include <numeric>#include <sstream>#include <stdexcept>#include <type_traits>#include <utility>#include <vector>#include “simulation.hpp”#include “/home/docs/checkouts/readthedocs.org/user_builds/moma/checkouts/add-docs–4/include/llg.hpp”#include “/home/docs/checkouts/readthedocs.org/user_builds/moma/checkouts/add-docs–4/include/integrators.hpp”#include “/home/docs/checkouts/readthedocs.org/user_builds/moma/checkouts/add-docs–4/include/io.hpp”#include “/home/docs/checkouts/readthedocs.org/user_builds/moma/checkouts/add-docs–4/include/field.hpp”#include “/home/docs/checkouts/readthedocs.org/user_builds/moma/checkouts/add-docs–4/include/trap.hpp”#include “/home/docs/checkouts/readthedocs.org/user_builds/moma/checkouts/add-docs–4/include/easylogging++.h”#include “/home/docs/checkouts/readthedocs.org/user_builds/moma/checkouts/add-docs–4/include/optimisation.hpp”#include “/home/docs/checkouts/readthedocs.org/user_builds/moma/checkouts/add-docs–4/include/dom.hpp”#include “/home/docs/checkouts/readthedocs.org/user_builds/moma/checkouts/add-docs–4/include/curry.hpp”#include <exception>#include <cblas.h>
Typedefs
- using
-
file
optimisation.hpp - #include <functional>#include <lapacke.h>
-
file
rng.hpp - #include <random>
-
file
simulation.hpp - #include <memory>#include <functional>#include <array>#include <random>#include <string>#include <cstdlib>#include <vector>#include “rng.hpp”#include “simulation.tpp”
Typedefs
- using
- using
-
file
stochastic_processes.hpp - #include <cstdlib>
-
file
trap.hpp - #include <cstdlib>
-
file
dom.cpp - #include ”../include/dom.hpp”#include ”../include/constants.hpp”#include ”../include/stochastic_processes.hpp”#include <cmath>
-
file
field.cpp - #include ”../include/field.hpp”#include <cmath>
Defines
-
_USE_MATH_DEFINES¶
-
-
file
integrators.cpp - #include ”../include/integrators.hpp”#include ”../include/optimisation.hpp”#include <cmath>
Typedefs
- using
- using
-
file
llg.cpp - #include ”../include/llg.hpp”#include ”../include/field.hpp”
-
file
moma_config.cpp - #include ”../include/moma_config.hpp”#include <cmath>#include <array>#include <string>#include <omp.h>#include <exception>#include ”../include/constants.hpp”#include ”../include/easylogging++.h”#include ”../include/field.hpp”#include ”../include/simulation.hpp”#include ”../include/rng.hpp”
Defines
-
_USE_MATH_DEFINES
Typedefs
- using
- using
-
-
file
optimisation.cpp - #include ”../include/optimisation.hpp”#include <cmath>#include <cblas.h>
-
file
rng.cpp - #include ”../include/rng.hpp”#include <stdexcept>
-
file
simulation.cpp - #include ”../include/simulation.hpp”
-
file
stochastic_processes.cpp - #include ”../include/stochastic_processes.hpp”#include <cblas.h>
-
file
trap.cpp - #include ”../include/trap.hpp”
-
file
README.md
-
file
main.cpp - #include <fstream>#include <array>#include <iostream>#include <random>#include <fenv.h>#include ”../include/moma_config.hpp”#include ”../include/json.hpp”#include ”../include/easylogging++.h”
Typedefs
- using
-
file
convergence.cpp - #include <random>#include <functional>#include <fstream>#include <cstdlib>#include ”../include/easylogging++.h”#include ”../include/moma_config.hpp”#include ”../include/simulation.hpp”#include ”../include/stochastic_processes.hpp”#include ”../include/io.hpp”
Functions
-
INITIALIZE_EASYLOGGINGPP int main(int argc, char * argv[])
-
-
file
equilibrium.cpp - #include <random>#include <functional>#include <fstream>#include <cstdlib>#include ”../include/easylogging++.h”#include ”../include/moma_config.hpp”#include ”../include/simulation.hpp”#include ”../include/stochastic_processes.hpp”#include ”../include/io.hpp”
Functions
-
INITIALIZE_EASYLOGGINGPP int main(int argc, char * argv[])
-
-
file
convergence.py
-
file
equilibrium.py
-
file
tests.cpp - #include ”../googletest/include/gtest/gtest.h”#include <limits>#include <ostream>#include <vector>#include “gtest/internal/gtest-internal.h”#include “gtest/internal/gtest-string.h”#include “gtest/gtest-death-test.h”#include “gtest/gtest-message.h”#include “gtest/gtest-param-test.h”#include “gtest/gtest-printers.h”#include “gtest/gtest_prod.h”#include “gtest/gtest-test-part.h”#include “gtest/gtest-typed-test.h”#include “gtest/gtest_pred_impl.h”#include ”../include/easylogging++.h”#include ”../include/llg.hpp”#include ”../include/integrators.hpp”#include ”../include/io.hpp”#include ”../include/simulation.hpp”#include ”../include/json.hpp”#include ”../include/moma_config.hpp”#include ”../include/trap.hpp”#include ”../include/optimisation.hpp”#include ”../include/rng.hpp”#include ”../include/stochastic_processes.hpp”#include ”../include/field.hpp”#include ”../include/dom.hpp”#include <cmath>#include <random>#include <lapacke.h>#include <stdexcept>
Functions
-
TEST(heun, multiplicative)¶
-
TEST(simulation, save_results)¶
-
TEST(heun_driver, ou)¶
-
TEST(trapezoidal_method, triangle)¶
-
TEST(moma_config, valid_llg_input)¶
-
TEST(moma_config, valid_dom_input)¶
-
TEST(moma_config, transform_llg)¶
-
TEST(moma_config, transform_dom)¶
-
TEST(newton_raphson, 1d_function)
-
TEST(newton_raphson, 1d_funtion_max_iter)
-
TEST(newton_raphson_noinv, 2d_function_2_sols)
-
TEST(newton_raphson_noinv, 2d_function_singular)
-
TEST(rng, mt_norm)¶
-
TEST(rng, array)¶
-
TEST(rng, array_stride_3)¶
-
TEST(rng, rng_mt_downsample)¶
-
TEST(implicit_integrator_midpoint, atest)¶
-
TEST(simulation, power_loss)¶
-
TEST(rk4, time_dependent_step)¶
-
TEST(master_equation, 3d_system)
-
-
page
todo
-
page
deprecated
-
dir
docs
-
dir
/home/docs/checkouts/readthedocs.org/user_builds/moma/checkouts/add-docs--4/include
-
dir
/home/docs/checkouts/readthedocs.org/user_builds/moma/checkouts/add-docs--4/lib
-
dir
/home/docs/checkouts/readthedocs.org/user_builds/moma/checkouts/add-docs--4/test/plotting
-
dir
/home/docs/checkouts/readthedocs.org/user_builds/moma/checkouts/add-docs--4/src
-
dir
/home/docs/checkouts/readthedocs.org/user_builds/moma/checkouts/add-docs--4/test
-
page
index A **M**odern **o**pen-source **m**agnetics simulation package.. **a**gain.
Simulate magnetic nano-particles with ease.
Features
- Stochastic Landau-Lifshitz-Gilbert equation
- Explicit Heun scheme integration
- OpenMP at the highest level. Never be ashamed of embarrassingly parallel
- Json for config files and output
- Real life logging!
- You won’t believe this is C++
Join the chat at:

Installation
Moma requires LAPACK and BLAS routines. Run the [makefile](makeflie) with intel complier:
``` shell $ make ```
Make with gcc (will need version >=4.9):
``` shell $ make CXX=g++-4.9 ```
Mac OSX
The easiest way to obtain LACPACK/BLAS is through homebrew by install openblas, which comes with both. Run the following command:
``` shell $ brew install homebrew/science/openblas ```
You’ll need to link to link to the libraries when running the make command, e.g:
``` shell $ make CXX=g++-4.9 LDFLAGS=-L/usr/local/opt/openblas/lib CXXFLAGS=-I/usr/local/opt/openblas/include ```
Tests
The fast unit tests can be run with
``` shell $ make tests $ ./test ```
The full test suite includes numerical simulations of convergence. These take a long time to execute (~5mins). Test suite results are available in
test/output.``` shell $ make run-tests $ cd test/output ```
Configuration
See the example config file for an overview of the options.
Dependencies
LAPACK: On Ubuntu:
``` shell $ sudo apt install liblapack-dev ```